Tools

With the awsome genetic power of Drosophila on our side, we develop and use cutting-edge techniques to investigate the mechanisms underlying the development and remodeling of neural circuits.
Tissue-specific CRISPR for large-scale in vivo screening
While genetic screens are a powerful tool for identifying novel genes involved in complex biological processes, traditional screening methods in Drosophila, including the popular RNAi strategy, hold various limitations. We have established and optimized a novel screening strategy, using a variation of the CRISPR/Cas9 technology, referred to as tissue-specific (ts)CRISPR. tsCRISPR enables precise targeting of any desired gene in a specific tissue or population of cells, within a single cross (10 days!). Being simple and fast, tsCRISPR is ideal for high-throughput in vivo screening. Using optimized tools, our lab has generated a large-scale resource of gRNA-expressing fly lines targeting hundreds of Drosophila genes, which is available to the worldwide fly community (https://bdsc.indiana.edu/stocks/genome_editing/sgrna.html). Our gRNA-collection can be readily used for large-scale screening in any Drosophila tissue or organ. In the mushroom body, it allowed the identification of many novel genes involved in neuronal remodeling.

RNA-seq at unprecedented temporal resolution
Both phases of mushroom body remodeling – pruning and regrowth – are regulated by nuclear receptors, suggesting that these processes are governed by genetic programs.
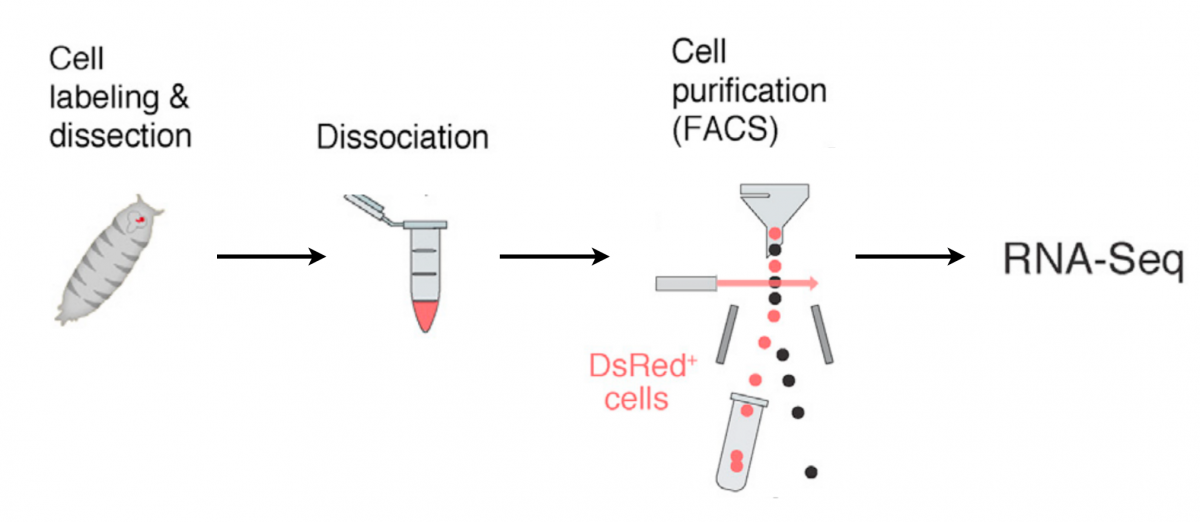
To uncover these programs, we recently isolated and profiled mushroom body neurons undergoing remodeling using FACS sorting followed by RNA-seq. Transcriptional profiling was performed at fine-grained temporal resolution – including every three hours during the remodeling period.
The transcriptional atlas we generated (see resources) provides invaluable insights about the molecular and cellular mechanisms that underlie neuronal remodeling.
Ex vivo brain culturing
A major hurdle in understanding the various dynamic processes that occur during neuronal remodeling is that they take place within the pupa, in which live-imaging is currently impossible (mainly due to opaque fat bodies that surround the brain). Therefore, we have developed an ex vivo long-term brain culturing system, which supports ongoing development of pupal brains, at similar kinetics to that which occurs in vivo. Using this new strategy, we have obtained the first time-lapse sequence of mushroom body remodeling in up to single cell resolution. Moreover, our culturing system enables pharmacological manipulations that provide important insights about the molecular basis of remodeling.

A sprouting assay of cultured neurons
The molecular mechanisms regulating intrinsic axon growth potential during development or following injury remain largely unknown, despite their fundamental significance. We have harnessed the genetic power of Drosophila to establish a neurite sprouting assay of primary cultured mushroom body neurons. Using this culturing system we can explore the sprouting characteristics of dissociated MB neurons – which mechanistically resembles axon regeneration after injury. By testing sprouting of WT and mutant MB neurons, this assay provides important insights about the intrinsic mechanisms of axon growth in both developmental and pathological contexts.

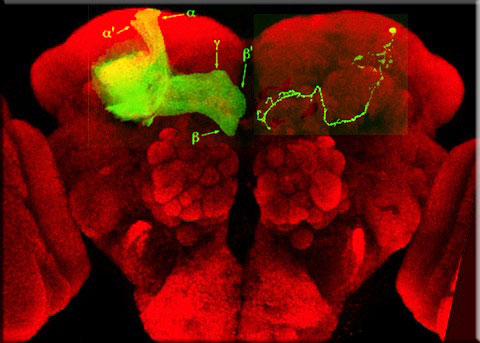
The MARCM technique for positive labeling of mutant clones
Lee & Luo 1999
A complex biological process occurring late in development, such as mushroom body remodeling, most likely utilizes molecules that are also required in a wide variety of other developmental processes. Mosaic analyses, which can be used to study such questions, was previously not useful in studying the nervous system where a single mutant axon needs to be visualized in the background of thousands of non-clonal axons. To solve this problem, the Luo laboratory developed the MARCM technique, which enables the creation of homozygous mutant clones (which can be as small as single cells), which are positively labeled. Using MARCM we can identify genes that are cell-autonomously required for mushroom body remodeling, as well as visualize structures within axons at subcellular resolution.